Hi,
I've put together a new script for outputting image statistics to delimited-format text files ready for processing in your favourite spread sheet or other data-munching application. The script is feature complete and this is the third release (no longer beta). The script is now part of the official PixInsight distribution so you should not need to install it manually if you are running the most recent version of PI and have installed all updates. The last version of the script plus installation instructions have been left in this post for reference only.
ChangeLog1.2 : Third (full) release).
Fixed script not aborting processing when dialog close button used.
Fixed re-sizing of file list so that scroll-bars appear when needed.
Relocated call to dialog.updateUI to resize exit button on launch.
1.1b: Second beta release.
Fixed incorrect restoration of settings from process icon.
Fixed problems with reentrant controls crashing script.
Added abort processing button/functionality.
Added PIDOC documentation and help button.
NoiseEvaluation-Engine.js out of beta.
ImageExtensions-lib.js out of beta.
GUIFactory-lib.js out of beta.
1.0b: First beta release.
Script Installation Instructions1. In the attached .zip file you'll find a folder called
"BatchStatistics". Place the unzipped folder and its contents in your
<PixInsight Folder>/src/scripts folder. (Note: Do not place the contents of the
"BatchStatistics" folder directly in to
/src/scripts but rather ensure that it is inside the
scripts folder in a sub-folder called
BatchStatistics).
2. In PixInsight select
"Script" menu ->
"Feature Scripts...".
3. Now click the
"Add" button from the
"Feature Scripts" dialog. Make sure the
<PixInsight Folder>/src folder is selected and click
"OK".
4. You should get a message saying that
"1 additional script(s) were found on directory ...". Click
"OK" and then
"Done".
5. The BatchStatistics Script should now appear under the
"Script" menu ->
"Batch Processing" ->
"BatchStatistics".
If you need more help, please see this video:
http://www.pixinsight.com.ar/en/docs/2/pixinsight-add-script.html.
Documentation Installation Instructions (Optional)7. To install the documentation unzip the
"pidoc" folder to a temporary location.
8. In PixInsight select
"Script" menu ->
"Development" ->
"Documentation Compiler".
9. Click
"Add Files" and browse to the pidoc folder that you just unzipped.
10. Choose the
"BatchStatistics.pidoc" file and click
"Open" to add it to the
"PIDoc Source Files" list.
11. Click the
"Run" button.
12. Click
"No" when asked if you want to perform another compilation.
13. The console should display
"PIDocCompiler: 1 succeeded, 0 failed, 17 warning(s)".
14. You can read the documentation by clicking the
"Documentation" button at the bottom left of the
"BatchStatistics" dialog, or by finding it in the
"Process Explorer" under
"<Scripts>" ->
"Batch Processing" ->
"BatchStatistics".
User Interface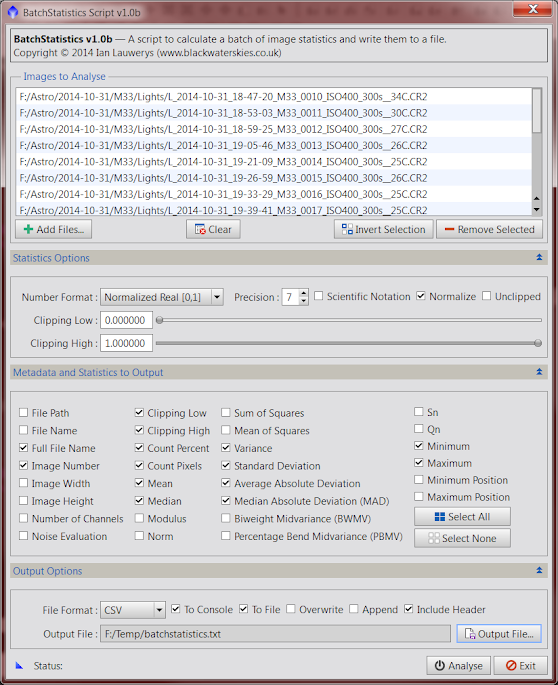 InstructionsImages to Analyse
InstructionsImages to Analyse1. Click the
"Add" button to choose image files to analyse. Any image file format supported by PixInsight should be suitable. The script will support images with multiple channels (RGB, etc.) and it
should also support multi-image file formats such as FITS. (Please note that his latter feature is untested by me so please report any problems via this thread).
Note that the dialog can be resized vertically to show more list entries and horizontally to show long file names by dragging its edges as required.
Important: When working with multi-channel images, it is best to try to work with sets of images that have the same number of channels and in the same order. BatchStatistics will process images with any number of channels, but will warn you via the PixInsight console if the current image contains a different number of channels than the previous one. Given that the objective is to output to a file format containing columns of data, changing the number of channels between images will mean that the column headers do not always correspond to the data rows. (Note that if you append results to an existing output file, BatchStatistics has no way to know how many columns of data it contains, so the operator is responsible for doing the right thing!)BatchStatistics does not currently support working with open image views, previews, etc. although I hope to add that functionality in a future version.
2. The
"Clear",
"Invert Selection" and
"Remove Selected" buttons function in the same manner as other PixInsight scripts and processes should you need to amend the list.
Statistics Options3. The
"Number Format" combo box functions in the same manner as the existing Statistics process. You may output results using PixInsight's native Normalized Real [0..1] format, or converted to Integer bit depth to match your camera's output format (typically 16 bit for CCD cameras or 12 / 14 bit for DSLR cameras).
4. The
"Precision" spin box allows you to choose the number of digits in Normalized Real and Scientific formats between 0 and 17 places.
5. The
"Scientific Notation" check box functions in the same manner as the existing Statistics process. It allows you to output Normalized Real format numbers in scientific notation.
6. The
"Normalize" check box functions in the same manner as the existing Statistics process. It makes all scale estimates consistent with the standard deviation of a normal distribution.
7. The
"Unclipped" check box functions in the same manner as the existing Statistics process. By default, statistics are computed by clipping (ignoring) pixels with values of 0 ("black") and 1 (fully saturated) when this box is unchecked. If you change the
"Clipping Low" and
"Clipping High" controls below, then pixels with values outside the chosen clipping range will be ignored instead. By checking the
"Unclipped" check box, statistics are computed for all pixels in the image ignoring any clipping range.
8.
"Clipping Low" and
"Clipping High": If the
"Unclipped" check box is unchecked, any pixels with values less than or equal to
"Clipping Low" or greater than or equal to
"Clipping High" will be excluded from statistics calculations.
Metadata and Statistics to Output9. Select the appropriate check boxes to choose which statistics to output. Except as noted below, all options should function in the same manner as the existing Statistics process.
Important: Selecting all statistics may lead to long pauses between images. The script may appear to have hung but be patient as some options (such as "Qn") are computationally expensive and may take some time to complete. The status bar and console messages will only update once all channels in an image have been fully processed.The following options are output once per image. The corresponding column headers are shown in
italics:
File Path: File_Path The path of the image being analysed.
File Name: File_Name The file name of the image being analysed.
Full File Name: File_Full_Name The combined path and file name of the image being analysed.
Image Number: Image_Number The image number within the file (useful for multi-image file formats only).
Clipping Low: Clipping_Low The low range clipping value for statistics calculations (Note: If "Unclipped" is selected, this column will be null).
Clipping High: Clipping_High The high range clipping value for statistics calculations (Note: If "Unclipped" is selected, this column will be null).
Image Width:
Width The width of the image in pixels.
Image Height: Height The height of the image in pixels.
Number of Channels: Channels_First_Analysed First channel number analysed.
Channels_Last_Analysed Last channel number analysed.
Channels_Analysed Number of channels analysed.
Note: that monochrome images usually have one channel (0), and colour images have three, Red (0), Green (1) and Blue (2), but this may vary depending on the image format.The following options are output once per channel in the image. The corresponding column headers are shown in
italics. Each column header will have the channel number to which it relates appended, e.g.
Mean_0,
Mean_1,
Mean_2, etc.
Count Percent:
Count_Pct_n Pixels used for statistics calculations (i.e. not clipped) as a percentage of total pixels in image.
Count Pixels:
Count_Px_n Number of pixels used for statistics calculations (i.e. not clipped).
Mean:
Mean_n The arithmetic mean, i.e. the average of sample values.
Median:
Median_n The median of sample values.
Modulus:
Modulus_n The sum of absolute sample values.
Norm:
Norm_n The sum of sample values.
Sum of Squares:
Sum_Of_Squares_n The sum of the squares of sample values.
Mean of Squares:
Mean_Of_Squares_n The mean of the squares of sample values.
Variance:
Variance_n The variance from the mean of sample values.
Standard Deviation:
StdDev_n The standard deviation from the mean of sample values.
Average Absolute Deviation:
AvgDev_n The average absolute deviation from the median of sample values.
Median Absolute Deviation (MAD):
MAD_n The median absolute deviation from the median (MAD) of sample values.
Biweight Midvariance (BWMV):
SQRT_BWMV_n The square root of the biweight midvariance of sample values.
Percentage Bend Midvariance (PBMV):
SQRT_PBMV_n The square root of the percentage bend midvariance of sample values.
Sn:
Sn_n The Sn scale estimator of Rousseeuw and Croux of sample values.
Qn:
Qn_n The Qn scale estimator of Rousseeuw and Croux of sample values.
Minimum:
Min_n The minimum sample value.
Maximum:
Max_n The maximum sample value.
Minimum Position:
Min_Pos_X_n The X coordinate of the first occurrence of the minimum sample value.
Min_Pos_Y_n The Y Coordinate of the first occurrence of the minimum sample value.
Maximum Position:
Max_Pos_X_n The X coordinate of the first occurrence of the maximum sample value.
Max_Pos_Y_n The Y Coordinate of the first occurrence of the maximum sample value.
The following additional measures are available over and above those in the existing Statistics process:
Noise Evaluation:
Sigma_MRS_n The noise standard deviation calculated using the Multi-resolution Support method.
Count_MRS_n The count of noise pixels calculated using the Multi-resolution Support method.
Layers_MRS_n The number of layers used by the Multi-resolution Support method.
Note: If the MRS method does not converge on a solution, these three columns will be null. Sigma_K_Sigma_n The noise standard deviation calculated using the K-Sigma method.
Count_K_Sigma_n The count of noise pixels calculated using the K-Sigma method.
Note: Noise Evaluation will produce the same results as the existing NoiseEvaluation script, except that the K-Sigma values will always be calculated regardless of whether the MRS method converges on a solution.10. The
"Select All" button checks all available statistics and metadata checkboxes.
Caution: Computing all statistics for a large number of images will take a significant amount of time, so only select those that you actually require.
11. The
"Select None" button unchecks all available statistics and metadata checkboxes.
Output Options12. The
"File Format" combo box allows you to select various delimited output formats from Tab, pipe, colon, space, comma or CSV.
Note: Any data value which contains the delimiter value will be enclosed in double quotes (" "). In the case of the CSV format, the delimiter is the comma character and all data values are enclosed in double quotes. Line endings are the MS-DOS CR LF format as per RFC 4180.13. The
"To Console" checkbox outputs header and result rows to the PixInsight Console. Due to informational messages (e.g. loading of images) this is not as useful as it could be as it is not possible to cut and paste a block of results directly from the console to your application, but it may be of use for single images (vs. cutting and pasting individual data values from the Statistics process).
Note: MS Excel's "Text to Columns" option is your friend when cutting and pasting from the console.14. The
"To File" checkbox outputs header and result rows to a text file.
15. The
"Overwrite" checkbox overwrites any existing text file of the same name as that specified in
"Output File".
Important: No warning will be given when overwriting (this is by design for future developments in re-using script instances) so please be careful!
16. The
"Append" checkbox appends results to any existing text file of the same name as that specified in
"Output File" or creates a new file if one does not exist.
17. The
"Include Header" checkbox outputs a row of column headers appropriate to the first image in the file list. Please see the note under 1. above about working with images containing varying numbers of channels.
Note: Headers are not written to existing files when appending, only to new ones if created (this is by design to ensure that each output file only contains a single header row at the top).18. The
"Output File..." button allows you to select a folder and select/specify a file name for output of results. The chosen file name is displayed in the
"Output File" text box.
Control Buttons19. The
"Analyse" button analyses all images in the file list and produces the selected statistics and metadata.
20. The
"Exit" button exits the BatchStatistics script.
Note: No warning is given upon exit so please take care to create a process icon if required.21. The blue
"Process" triangle can be dragged to the PixInsight workspace to create a process icon. All file entries in the list and the options are saved as part of the process icon. The process may be re-instantiated by right clicking the script icon, choosing
"Launch Script Instance" and then clicking the round
"Apply Global" icon at the bottom of the
"Script" dialog.
Note: Dragging the "Process" triangle on to an image window has no effect at present. It is planned to allow processing of image statistics from open images using this method in a future version.22. The blue
"Documentation" button can be clicked to browse the script documentation (if you installed it), or hovered over for brief instructions.
Notes for Script Developersa. The "ImageExtensions-lib.js" file contains methods to extend the PJSR Image.prototype to add .count() and .variance() methods, which are available in PCL and also in the (out of date) PJSR ImageStatistics object, but not available on the PJSR Image object. This is code supplied to me by Juan, all I have done is added a bit of wrapper around it to check for the existence of these methods and any conflicting property names. Juan said that he will add these methods in a future release, so the wrapper should ensure there is no conflict as and when that happens. As ever feel free to re-use per the license in the file - it's not my code so no credit claimed!
b. The "NoiseEvaluation-Engine.js" file duplicates the functionality of the NoiseEvaluation script but in a form that can be re-used by other scripts. The code is documented so if you need these measures in your own code feel free to re-use, again respecting the license and again acknowledging that this is not my code, just a refactoring of Juan's.
c. The "BatchStatistics-Engine.js" file allows (reasonably) efficient processing of image files to calculate statistics. The data can be accessed in delimited text format or directly from the public properties of the StatisticsEngine object. All methods and properties are documented in the code. I plan to extend this object in the future to re-use in other scripts (I have various ideas). Again feel free to re-use respecting the license details.
d. The "GUIFactory-lib.js" contains a rough and ready factory object to simplify creation of UI controls in scripts. It is far from feature-complete and really designed to save me time typing and constantly looking up other examples (there are a
lot of controls in my little script!). Again feel free to re-use or fork as required. Others have done similar things in their scripts, so I'm not looking for any prizes for originality here!
As a general note, lots of people create scripts with separation between processing engines and the GUI, but unfortunately they are often wrapped up in a single script file. It might be going a bit far to suggest some kind of official script-library, but I think it would encourage development if we did try to split scripts in to separate component files with re-usable elements as some already do.